Distinct passing profiles in Statsbomb 360 data

The concepts in this post are extensively borrowed from Andy Rowlinson’s work.
Isolation Forest algorithm for Statsbomb 360 data
Statsbomb have released Women’s Euro 2022 event data alongside their Men’s Euro 2020 data, presenting a good opportunity for some Machine Learning analysis on a rich event dataset.
Andy Rowlinson demonstrated an Isolation Forest algorithm using Euro 2022 passing data, as a method to answer the question: “which players have the most unusual passing profiles”. This blog reimplements that approach using R.
What does an Isolation Forest do?
An Isolation Forest algorithm identifies anomalies in a dataset. The image below shows anomaly scores for a two-dimensional dataset, with greater anomalies in darker red.
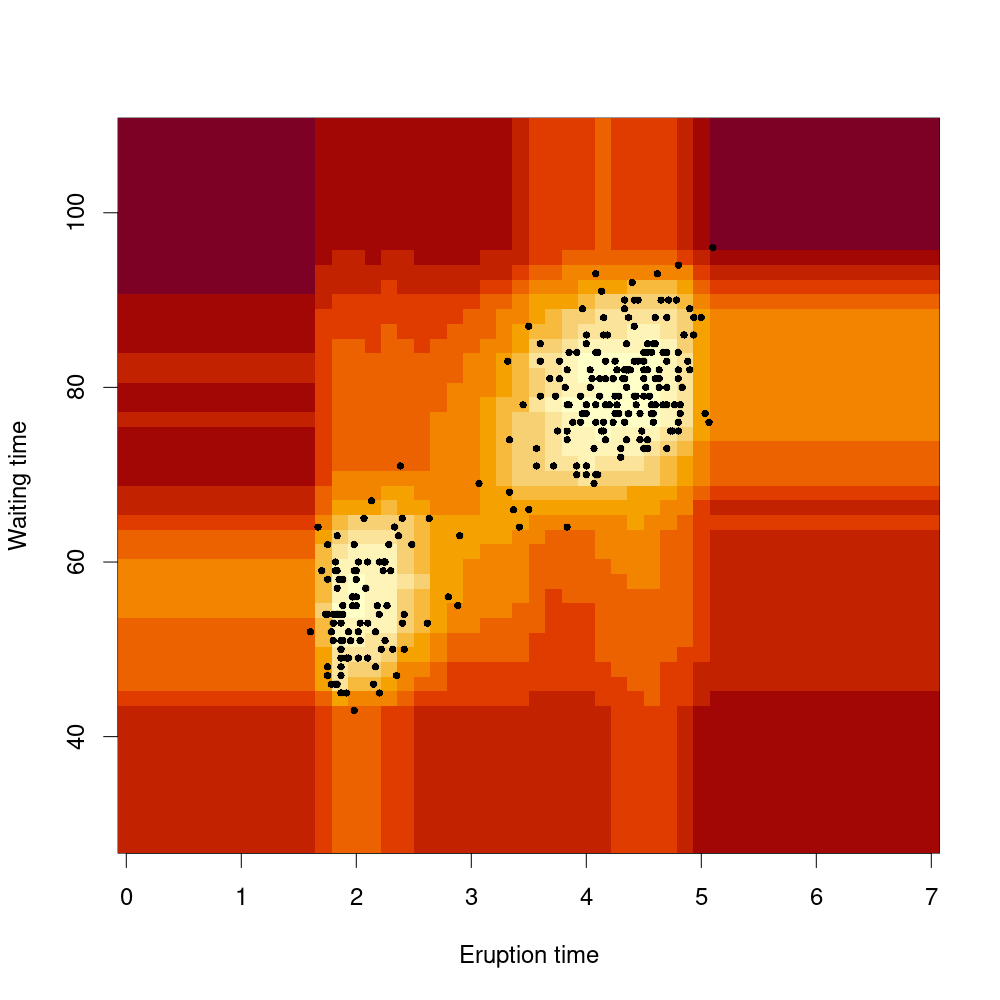
An Isolation Forest generalises this to identify outliers in data with many dimensions.
Packages
library(tidyverse)
library(StatsBombR)
library(ggsoccer)
library(isotree)
StatsBombR package installation guide
The isotree R package is used to create the Isolation Forest model, because it is the most useable model I can find that isn’t specifically designed for time series data.
Data import
FreeCompetitions <- FreeCompetitions()
Euro2022 <- FreeCompetitions %>%
filter(competition_id==53)
Euro2022Matches <- FreeMatches(Euro2022)
Euro2022Events <- free_allevents(MatchesDF=Euro2022Matches, Parallel=T) %>% allclean()
Import following the StatsBomb Working with R guide.
Pre-processing
passes <-
Euro2022Events %>%
# filter pass events
filter(type.name %in% c("Pass")) %>%
# remove set piece pass types
filter(!(pass.type.name %in% c("Kick Off","Goal Kick","Corner","Throw-in","Free Kick"))) %>%
# keep player name and pass location
select(
player.name,
location.x,location.y,
pass.end_location.x,pass.end_location.y
) %>%
# bin into pitch zones
mutate(
# treat the defensive half as one zone
# split the attacking half into 20 x 20 zones
across(where(is.numeric) & contains("x"),cut,breaks=c(0,60,80,100,120),.names="{.col}.bin"),
across(where(is.numeric) & contains("y"),cut,breaks=seq(0,80,20),.names="{.col}.bin"),
across(contains(".bin"),as.character)
) %>%
mutate(
# single y axis zone for the defensive half
location.y.bin=ifelse(location.x.bin=="(0,60]","(0,80]",location.y.bin),
pass.end_location.y.bin=ifelse(pass.end_location.x.bin=="(0,60]","(0,80]",pass.end_location.y.bin),
)
player.name location.y location.y.bin
<chr> <dbl> <chr>
1 Sarah Puntigam 45.2 (0,80]
2 Viktoria Schnaderbeck 37.6 (0,80]
3 Carina Wenninger 66.7 (0,80]
4 Rachel Daly 13.6 (0,80]
5 Sarah Zadrazil 63.4 (60,80]
6 Nicole Billa 57.1 (40,60]
7 Barbara Dunst 51.5 (40,60]
8 Laura Feiersinger 37.8 (20,40]
(location.y and location.y.bin columns only)
This creates a data frame of passes containing: player name, pass start location and pass end location.
In StatsBomb data, the pitch has dimensions of 120x80. The Isolation Forest requires categorised data, so the start and end locations are binned into:
- A 20x20 grid in the attacking half (4 squares wide and 3 squares long)
- A single bin in the defensive half
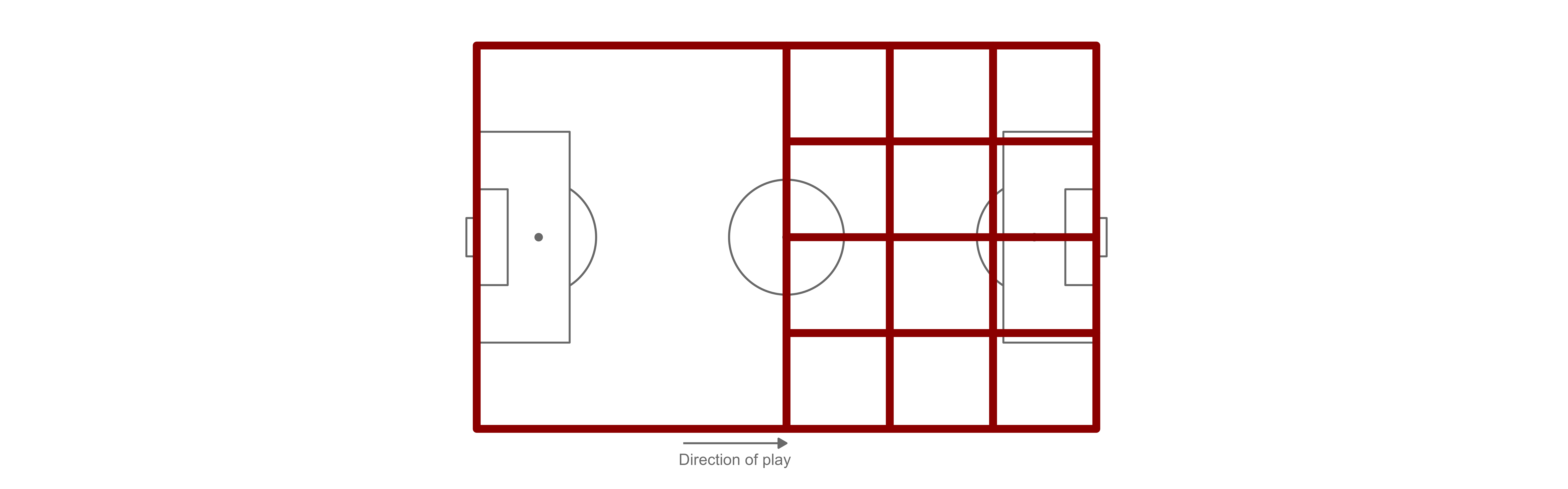
# count the total number of passes by each player
passes_total <-
passes %>%
group_by(player.name) %>%
summarise(passes=n(),.groups="drop") %>%
arrange(desc(passes))
player.name passes
<chr> <int>
1 Leah Williamson 522
2 María Pilar León Cebrián 393
3 Keira Walsh 359
4 Irene Paredes Hernandez 351
5 Millie Bright 347
Passing totals are useful to have for later.
Transition matrix
transition_matrix <-
passes %>%
group_by(player.name,across(contains(".bin"))) %>%
summarise(passes=n(),.groups="drop") %>%
pivot_wider(
names_from = c(location.x.bin,location.y.bin,pass.end_location.x.bin,pass.end_location.y.bin),
names_sort = TRUE,
values_from = passes,
values_fill = 0
)
Rows: 308
Columns: 146
$ player.name <chr> "Abbie Magee", "Ada Stolsmo Hegerberg", "Adelina Engman", "Agla Marí…
$ `(0,60]_(0,80]_(0,60]_(0,80]` <int> 15, 12, 7, 7, 6, 70, 70, 7, 48, 5, 17, 1, 20, 93, 0, 11, 1, 13, 19, …
$ `(0,60]_(0,80]_(100,120]_(0,20]` <int> 0, 0, 0, 0, 0, 0, 0, 0, 2, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 2,…
$ `(0,60]_(0,80]_(100,120]_(20,40]` <int> 0, 0, 0, 0, 0, 0, 0, 0, 2, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1,…
$ `(0,60]_(0,80]_(100,120]_(40,60]` <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 2, 0, 0, 0, 0, 0, 0, 0, 0, 1,…
$ `(0,60]_(0,80]_(100,120]_(60,80]` <int> 1, 0, 0, 0, 0, 2, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ `(0,60]_(0,80]_(60,80]_(0,20]` <int> 0, 1, 0, 1, 0, 1, 4, 0, 5, 0, 3, 0, 0, 1, 0, 2, 0, 0, 0, 4, 4, 0, 7,…
(matrix transposed with glimpse - player names are the rows, columns are coordinates in x_y_xend_yend format. Matrix view.
This section creates a data frame transition_matrix of players (308 rows) and start/end pass zones (146 columns), counting the proportion of passes starting and ending in each zone.
Many passes begin and end in the defensive half, represented by the (0,60]_(0,80]_(0,60]_(0,80] category.
transition_matrix_normal <-
transition_matrix %>%
# join total passes column
inner_join(passes_total) %>%
# divide by total player passes to normalise rows
mutate(across(!c(player.name,passes),~.x/passes))
Rows: 308
Columns: 146
$ player.name <chr> "Abbie Magee", "Ada Stolsmo Hegerberg", "Adelina Engman", "Agla Marí…
$ `(0,60]_(0,80]_(0,60]_(0,80]` <dbl> 0.484, 0.207, 0.318, 0.318, 0.333, 0.737, 0.235, 0.156, 0.338, 0.556…
$ `(0,60]_(0,80]_(100,120]_(0,20]` <dbl> 0.000, 0.000, 0.000, 0.000, 0.000, 0.000, 0.000, 0.000, 0.014, 0.000…
$ `(0,60]_(0,80]_(100,120]_(20,40]` <dbl> 0.000, 0.000, 0.000, 0.000, 0.000, 0.000, 0.000, 0.000, 0.014, 0.000…
$ `(0,60]_(0,80]_(100,120]_(40,60]` <dbl> 0.000, 0.000, 0.000, 0.000, 0.000, 0.000, 0.000, 0.000, 0.000, 0.000…
$ `(0,60]_(0,80]_(100,120]_(60,80]` <dbl> 0.032, 0.000, 0.000, 0.000, 0.000, 0.021, 0.000, 0.022, 0.000, 0.000…
$ `(0,60]_(0,80]_(60,80]_(0,20]` <dbl> 0.000, 0.017, 0.000, 0.045, 0.000, 0.011, 0.013, 0.000, 0.035, 0.000…
(matrix transposed with glimpse - rows are player names, columns are coordinates in x_y_xend_yend format) Matrix view.
The data is normalised to the interval [0,1] by dividing by the total number of passes per player. This ensures that total pass volume does not effect whether a pass profile is distinct.
Isolation Forest training
# create isolation forest using isotree function
forest <-
transition_matrix_normal %>%
select(-player.name,-passes) %>%
isolation.forest(
data=.,
ndim=1,
ntree=500,
missing_action="fail",
scoring_metric="density"
)
The Isolation Forest is trained on the transition matrix. 500 trees is sufficient to give repeatable results.
I have replicated the approach in the isotree Isolation Forest guide to train this model, mostly the example with real data which trains the Isolation Forest on a large matrix.
Prediction
# calculate the isolation prediction of each player
prediction <-
# join the prediction to the transition matrix containing player names
bind_cols(
# predict using isotree function
predict(forest, transition_matrix_normal) %>%
tibble("anomaly_score"=.),
# join to transition matrix
transition_matrix_normal
) %>%
# keep the player name and prediction
select(player.name,anomaly_score) %>%
# join the player passes total
inner_join(passes_total) %>%
# only keep players with at least 20 passes
filter(passes >= 20) %>%
# order by prediction
arrange(desc(anomaly_score))
player.name anomaly_score passes
<chr> <dbl> <int>
1 Francisca Ramos Ribeiro Nazareth Sousa -4.78 31
2 Aitana Bonmati Conca -5.06 298
3 Jill Roord -5.09 134
4 Ella Toone -5.23 130
5 Barbara Bonansea -5.25 56
6 Andreia Alexandra Norton -5.37 98
7 Bethany Mead -5.40 145
8 Charlotte Bilbault -5.40 187
9 Nicole Billa -5.45 80
10 María Francesca Caldentey Oliver -5.56 273
11 Cristiana Girelli -5.56 58
12 Lea Schüller -5.67 29
The anomaly score of each player pass profile is calculated. The isotree::predict function creates an array of predictions which corresponds to the players in the isolatioin matrix, joined back to the matrix with bind_cols.
Players with higher (closer to zero) anomaly_score are the most isolated profiles identified by the algorithm.
Results
# inner join to only keep player data from the top 12 predictions
inner_join(
passes,
prediction %>%
slice_max(anomaly_score,n=12)
) %>%
# wrap longer names
mutate(player.name=str_wrap(player.name,width=25)) %>%
# order by ranking
mutate(player.name=fct_reorder(player.name,desc(anomaly_score))) %>%
ggplot() +
annotate_pitch(dimensions=pitch_statsbomb) +
geom_segment(aes(x=location.x,xend=pass.end_location.x,y=location.y,yend=pass.end_location.y),size=0.3,arrow=arrow(length=unit(0.1, "cm"))) +
theme_pitch() +
theme(
strip.text=element_text(size=6),
strip.background=element_blank(),
plot.caption=element_text(face="italic",size=6)
) +
facet_wrap(vars(player.name),ncol=4) +
labs(
title="Most unique Euro 2022 passing profiles",
caption="Statsbomb 360 data"
)
Congratulations to Francisca Nazareth of Portugal, whose 31 passes were more distinct than some more well-known creative passers such as Aitana Bonmatí (pictured above).
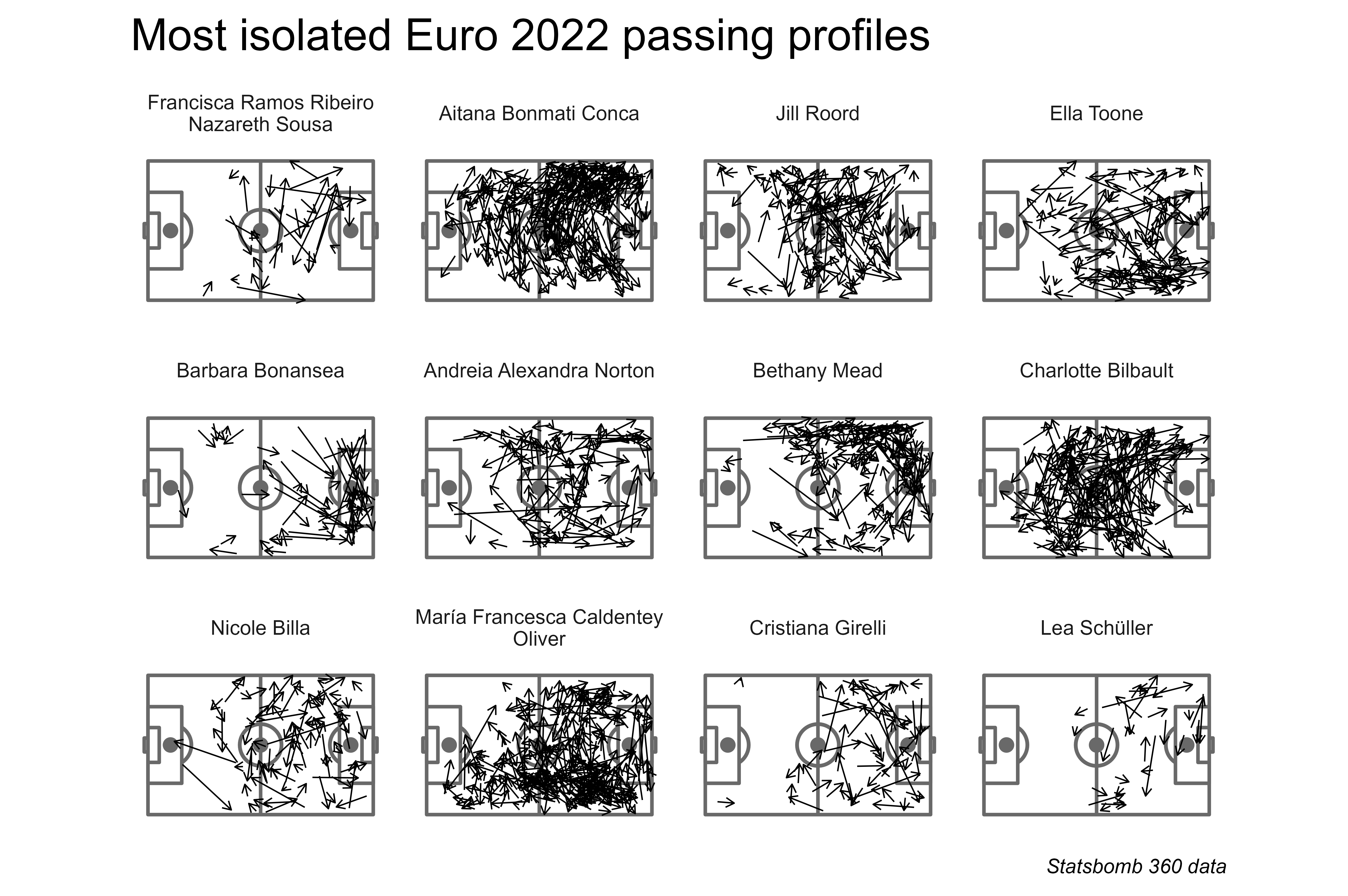
Three players identified in this implementation also appeared in Andy’s implementation:
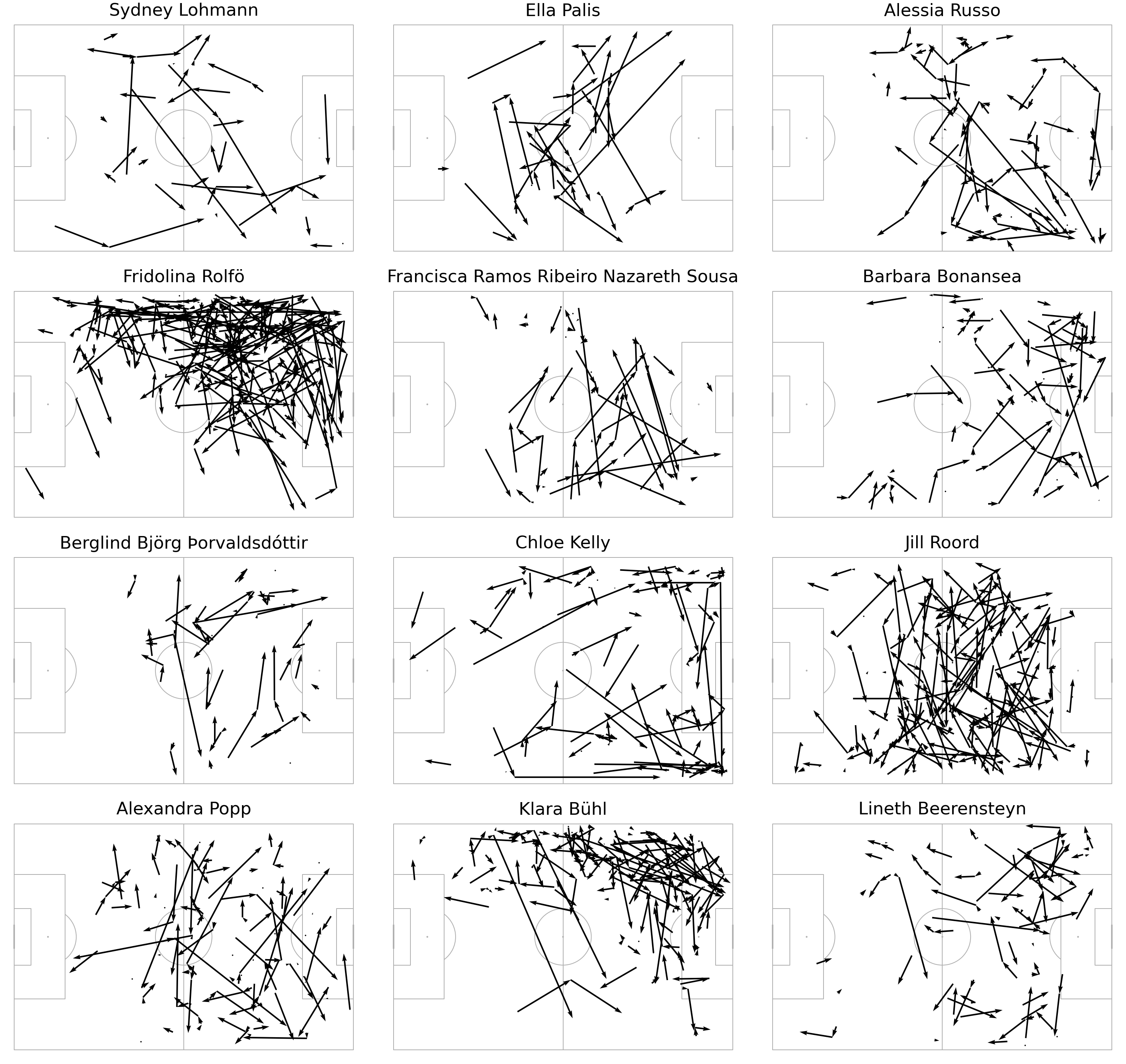
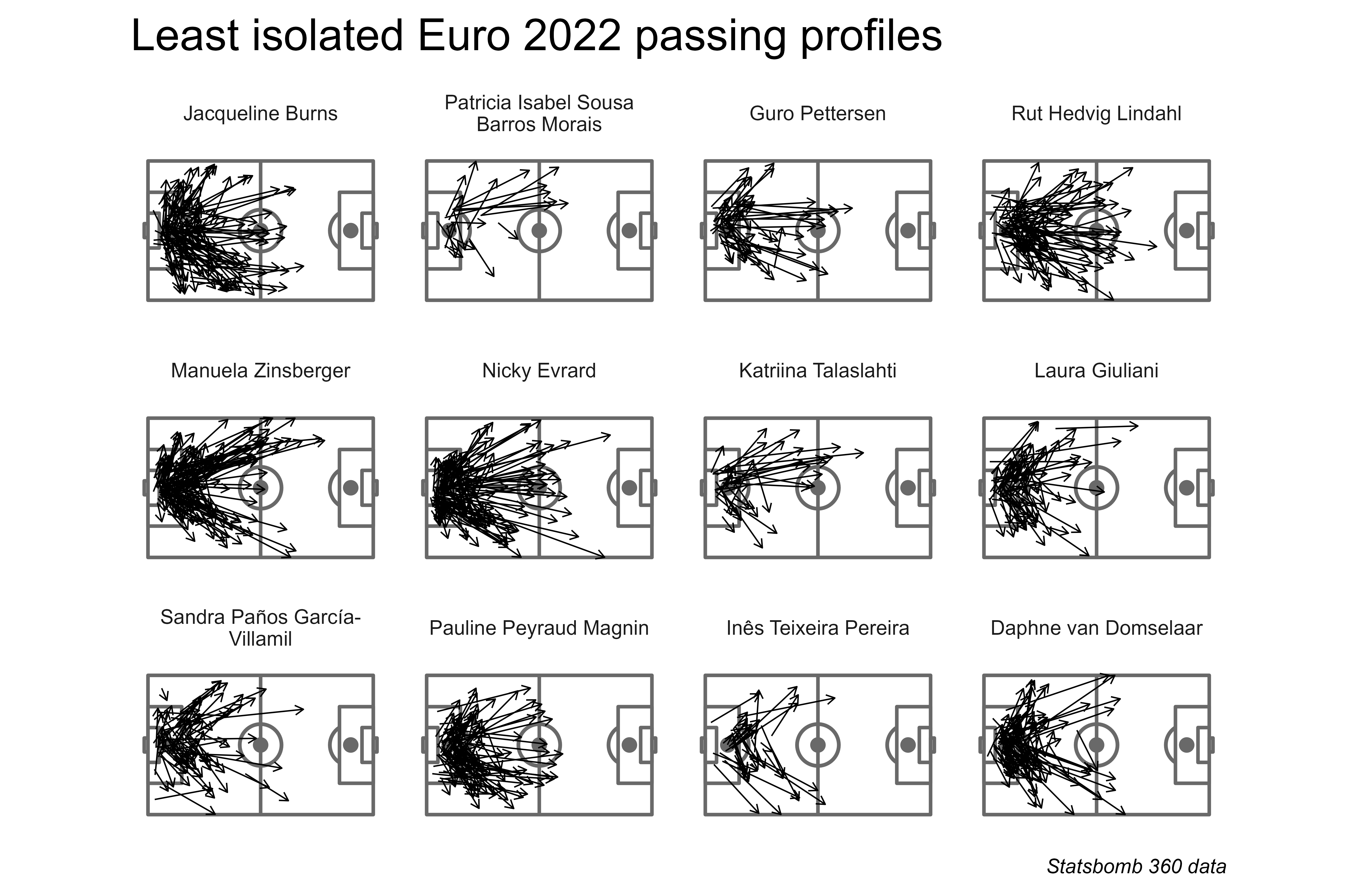
Naturally, the least isolated set of players were mostly goalkeepers. Grouping the defensive half as a single zone helps to differentiate attacking passers over defensive ones.
What about testing the algorithm with Men’s Euro 2020 data?
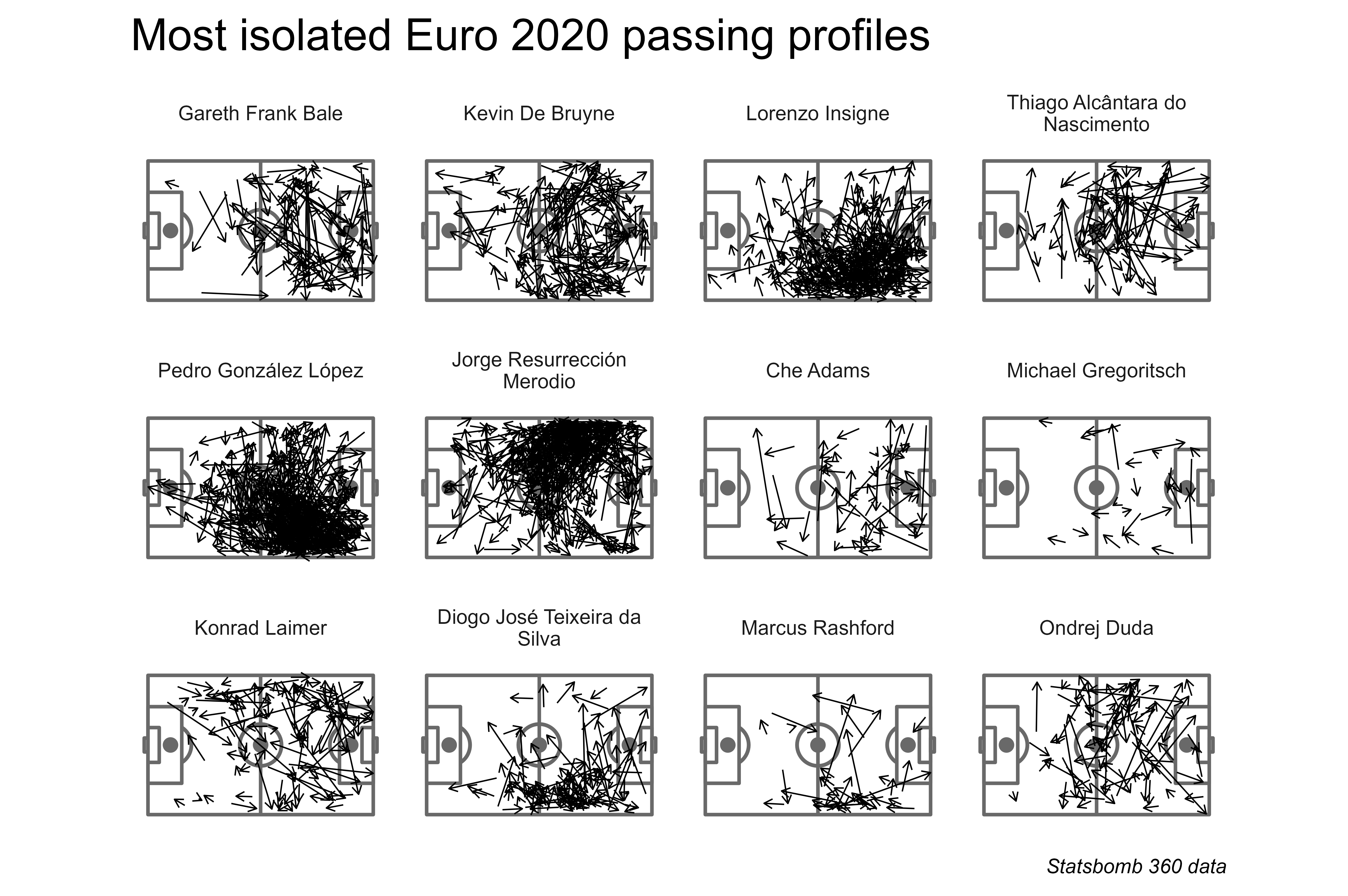
It’s notable that some players who carried a lot of their country’s passing have the most distinct passing profiles with this algorithm.
Limitations
- Players who didn’t progress through the knockout stages have limited passing data.
- Distinct passing profile may not indicate actual passing ability.
- Zones are discrete features and the model doesn’t know which zones are adjacent.
- Passing location is affected by teams playing higher or lower lines.
- Players who can play in multiple positions (such as on both wings) are over-identified, even if their actual passing characteristics are typical for wide players.
- Midfield passers from just inside the attacking half may be over-identified.
- Progressive passers from some deep positions may be under-identified.
- Passers into very dangerous penalty area positions may be under-identified.
Thanks to
- Andy Rowlinson for sharing his original work and allowing me to borrow his ideas to reproduce in R (mistakes are mine).
- StatsBomb for providing Euro 2022 and Euro 2020 event data.
- David Cortes, creator of
isotree. - Ben Torvaney, creator of
ggsoccer.
Questions, comments and complaints to:
- saintsbynumbers@gmail.com
- @saintsbynumbers on Twitter
- szfh on Github